Abstract
Background
Myelodysplastic syndromes (MDS) are clonal stem cell malignancies characterized by cytopenia, inefficient hematopoiesis, dysplasia in one or more myeloid cell lineages and increased risk of development of acute myeloid leukemia. It has been well appreciated that genomic alterations play a key role in MDS pathogenesis. The Revised International Prognostic Scoring System (IPSS-R) algorithm is commonly used to predict overall survival but may fail to recapitulate reliable prognostic information at the individual patient level, especially at the time of hematopoietic cell transplantation (HCT). Current World Health Organization (WHO) classification includes MDS with isolated 5q deletion as the only genetically defined category. Comprehensive analysis of recurrent genomic features by unsupervised clustering empowers discovery of potential prognostic molecular signatures.
Methods
Using whole blood samples obtained from 494 MDS patients at the time of HCT, we conducted whole-genome sequencing (WGS) and somatic variant processing via a custom analytic pipeline based on OCTOPUS and a set of annotation databases. Multiple filters allowed for selection and fine-tuning of criteria, including removal of variants with Gnomad allele frequency above 10x10 -06, removal of noncoding variants in low complexity and repetitive regions, those with no functional indications from ANNOVAR annotations, CADD conservative score under 15, and absence in HGMD or COSMIC databases. Highly annotated clinical data, including cytogenetic abnormalities at the latest time point prior to HCT, were obtained from CIBMTR forms. K-means clustering was applied to recurrent mutations and cytogenetic abnormalities to identify clinically relevant genomic subtypes. The optimal cluster number was determined by Gap-status algorithm. Statistics of clinical characteristics were compared among different genomic subgroups by Chi-squared test for categorical variables and Mann-Whitney U test for continuous variables. Overall survival association tests were conducted by Cox multivariate models. Relapse and transplant-related risk were performed by competing risk analysis using Fine-Gray models. Models were adjusted for patient-, disease-, and HCT-related factors.
Results
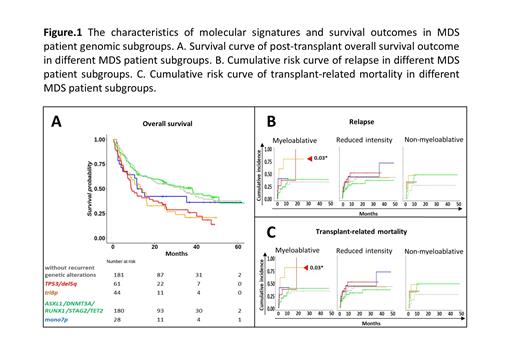
The somatic genomic landscape in our MDS cohort was examined for the total count of recurrent mutations at the sample level and gene level. Among 53 recurrently mutated genes in 257 of 494 MDS cases, TP53, TET2, RUNX1, DNMT3A, and ASXL1 were the most frequently mutated genes in our MDS cohort. Based on k-means clustering of the recurrent mutational and cytogenetic data, we detected five clusters that stratified our MDS patient cohort, including one reference cluster with no recurrent somatic mutations or cytogenetic abnormalities. Compared to the reference subgroup, significantly higher cytogenetic scores and IPSS-R scores were observed in genomic clusters with TP53 mutations (cytogenetic score: P=3.42E-07*; IPSS-R score: P= 2.38E-10*) and cytogenetic abnormalities del5q, or tri8p (cytogenetic score: P= 2.38E-10*; IPSS-R score: P=0.09) , or mono7 (cytogenetic score: P=3.29E-13*; IPSS-R score: P=1.38E-05*) (data not shown). Cox multivariate models revealed that genomic clusters with TP53 and del5q mutations (P<0.001*) or tri8p (P=0.02*) mutations have strong associations with post-transplant overall survival outcome (Figure 1A). Furthermore, competing risk analysis confirmed significantly higher risk of relapse in genomic subgroups with TP53 and del5q mutations in the reduced intensity conditioning regimen setting (P=0.01) (Figure 1B), while significantly higher risk of transplant-related mortality was found in the genomic subgroup with tri8p in the myeloablative conditioning regimen setting (P=0.03) (Figure 1C).
Conclusion
Our study suggests that molecular signatures from MDS patient genomes at HCT may provide an independent prognosis of post-transplant survival. Additionally, our data suggests that the choice of regimen intensity could be informed by knowledge of the individual genomic signature of a given MDS patient.
Saber: Govt. COI: Other.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal